
Evaluate (and possibly plot) the General Dynamic Response Function (GDRF) for an autoregressive distributed lag (ADL) model
Source:R/tseffects.R
GDRF.adl.plot.RdEvaluate (and possibly plot) the General Dynamic Response Function (GDRF) for an autoregressive distributed lag (ADL) model
Usage
GDRF.adl.plot(
model = NULL,
x.vrbl = NULL,
y.vrbl = NULL,
d.x = NULL,
d.y = NULL,
shock.history = "pulse",
inferences.y = "levels",
inferences.x = "levels",
dM.level = 0.95,
s.limit = 20,
se.type = "const",
return.data = FALSE,
return.plot = TRUE,
return.formulae = FALSE,
...
)Arguments
- model
the
lmmodel containing the ADL estimates- x.vrbl
named vector of the x variables and corresponding lag orders in the ADL model
- y.vrbl
named vector of the (lagged) y variables and corresponding lag orders in the ADL model
- d.x
the order of differencing of the x variable in the ADL model
- d.y
the order of differencing of the y variable in the ADL model
- shock.history
the desired shock history.
shock.historydetermines the shock history (h) that will be applied to the independent variable. -1 represents a pulse. 0 represents a step. These can also be specified viapulseandstep. For others, see Vande Kamp, Jordan, and Rajan. The default ispulse- inferences.y
does the user want resulting inferences about the dependent variable in levels or in differences? (For y variables where
d.yis 0, this is automatically levels.) The default islevels- inferences.x
does the user want to apply the shock history to the independent variable in levels or in differences? (For x variables where
d.xis 0, this is automatically levels.) The default islevels- dM.level
significance level of the GDRF, calculated by the delta method. The default is 0.95
- s.limit
an integer for the number of periods to determine the GDRF (beginning at s = 0)
- se.type
the type of standard error to extract from the model. The default is
const, but any argument tovcovHCfrom thesandwichpackage is accepted- return.data
return the raw calculated GDRFs as a list element under
estimates. The default isFALSE- return.plot
return the visualized GDRFs as a list element under
plot. The default isTRUE- return.formulae
return the formulae for the GDRFs as a list element under
formulae(for the GDRFs) andbinomials(for the shock history). The default isFALSE- ...
other arguments to be passed to the call to plot
Examples
# ADL(1,1)
# Use the toy data to run an ADL. No argument is made this is well specified; it is just expository
model.toydata <- lm(y ~ l_1_y + x + l_1_x, data = toy.ts.interaction.data)
# Pulse effect of x
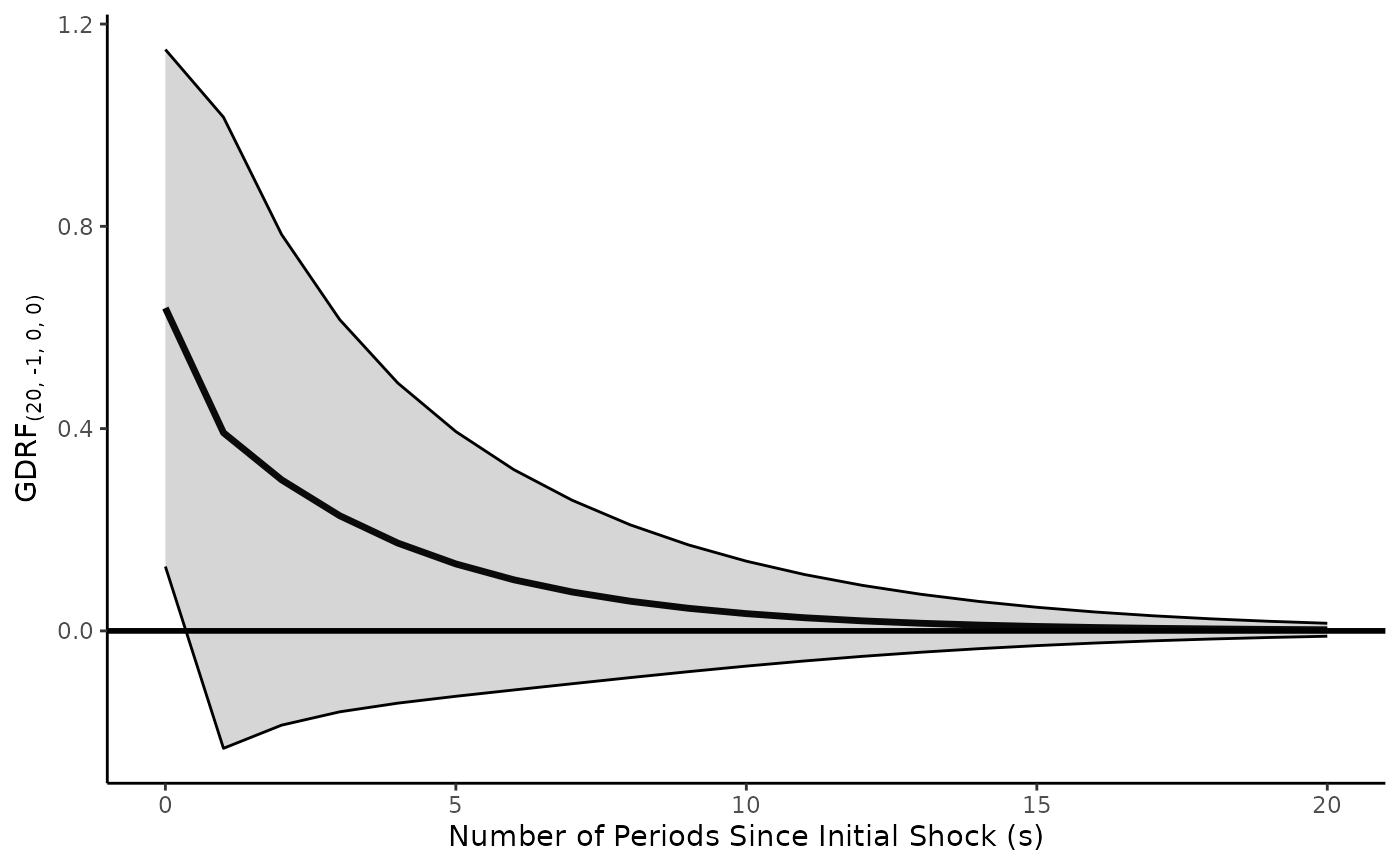
GDRF.adl.plot(model = model.toydata,
x.vrbl = c("x" = 0, "l_1_x" = 1),
y.vrbl = c("l_1_y" = 1),
d.x = 0,
d.y = 0,
shock.history = "pulse",
inferences.y = "levels",
inferences.x = "levels",
s.limit = 20)
 # Step effect of x. You can store the data to draw your own plot,
# if you prefer
test.cumulative <- GDRF.adl.plot(model = model.toydata,
x.vrbl = c("x" = 0, "l_1_x" = 1),
y.vrbl = c("l_1_y" = 1),
d.x = 0,
d.y = 0,
shock.history = "step",
inferences.y = "levels",
inferences.x = "levels",
s.limit = 20)
test.cumulative$plot
#> NULL
# Step effect of x. You can store the data to draw your own plot,
# if you prefer
test.cumulative <- GDRF.adl.plot(model = model.toydata,
x.vrbl = c("x" = 0, "l_1_x" = 1),
y.vrbl = c("l_1_y" = 1),
d.x = 0,
d.y = 0,
shock.history = "step",
inferences.y = "levels",
inferences.x = "levels",
s.limit = 20)
test.cumulative$plot
#> NULL